Description
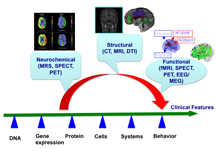
The term “Neuroimaging Data Analysis” (NDA) is aimed at encompassing a broad array of imaging, mathematical, and statistical methods for the analysis of neuroimaging data. This NDA program is motivated by the great need in the analysis of high-dimensional, correlated, and complex neuroimaging data and clinical and genetic data obtained from various cross-sectional and clustered neuroimaging studies. These neuroimaging studies usually collect structural, neurochemical, and functional images as well as clinical and genetic data. See Figures 1 and 2 for an illustration.
These structural, neurochemical, and functional imaging modalities include computed axial tomography (CT), diffusion tensor imaging (DTI), functional magnetic resonance imaging (fMRI), magnetic resonance imaging (MRI), magnetic resonance spectroscopy (MRS), positron emission tomography (PET), single photon emission tomography (SPECT), electroencephalography (EEG), and magnetoencephalography (EMG), among many others. The NDA program seeks to bring together a diverse group of researchers from different disciplines including statistics, mathematics, computer science, biomedical engineering, psychiatry, psychology, neuroscience, radiology, and beyond, to explore the common structure that underlies NDA methodologies, with a view toward motivating and synthesizing new approaches.
Research foci
- Image reconstruction
- Image segmentation
- Image registration
- Shape analysis
- Statistical group analysis
- Connectivity analysis
- Multimodal analysis
- Imaging genetics
- NDA applications
Schedule of activities
This two-week program ran from 4 – 14 June 2013 at SAMSI in Research Triangle Park.
The first four days, from Tuesday 4 June to Saturday 7 June, was spent on training courses concerning structural and functional neuroimaging data analysis, taught by leading researchers, to bring everyone up to speed on currently used methodology.
The second week, from Sunday 9 June to Friday 14 June, combined working groups (in the afternoons) with a workshop (in the mornings). Twenty invited distinguished speakers addressed major areas and cutting edge research in NDA. There was a poster session on Monday 10 June to allow all participants to present their ideas.
There were four working groups, with the schedule arranged so that participants had the option to join more than one group.
Training courses
The four-day short course on structural and functional neuroimaging data analyses from 4 – 7 June had the following planned instructors:
- John Aston, Warwick (Statistics); lecture notes
- F. DuBois Bowman, Emory (Biostatistics); lecture notes
- Martin Lindquist, Johns Hopkins (Biostatistics); lecture notes
- Marc Niethammer, UNC (Computer Science); lecture notes
- Hernando C. Ombao, UC Irvine (Statistics)
- Dinggang Shen, UNC (Radiology)
- Martin Styner, UNC (Computer Science and Psychiatry; lecture notes
- Hongtu Zhu, UNC (Biostatistics); lecture notes_1; lecture notes_2
Content: The lecture contents included the major scientific questions, mathematical and statistical methods, and their applications. The first two days was focused on structural NDA. Specifically, they covered statistical and mathematical methods associated with imaging reconstruction, imaging segmentation, imaging registration, and their applications. The last two days focused on functional NDA, including statistical and mathematical methods associated with imaging reconstruction, group analysis, and their applications.
Prerequisites: Basic knowledge of multivariate calculus, differential equation, and linear regression.
Working Groups
Discussions on cutting-edge research topics were divided into four working groups:
W1. Group Analysis (Chairs: Hongtu Zhu, UNC-CH and Tingting Zhang, Virginia)
W2. Imaging Registration (Chairs: Steven Marron, UNC-CH and Marc Niethammer, UNC-CH)
W3. Connectivity Analysis (Chairs: Daniel Rowes, Marquette and Jing Zhang, Yale)
W4. Multimodal Analysis (Chairs: Ombao Hernando, UC Irvine and Martin Lindquist, Johns Hopkins)
The goal of these working groups was to stimulate collaborations between researchers in the neuroimaging, mathematical, and statistical communities. Each working group discussed several important issues including existing approaches and their limitations, cutting-edge research questions, important mathematical and statistical methods for solving those questions, and collaborations among group members.