2014-15 Program on Beyond Bioinformatics: Statistical and Mathematical Challenges (Bioinformatics)
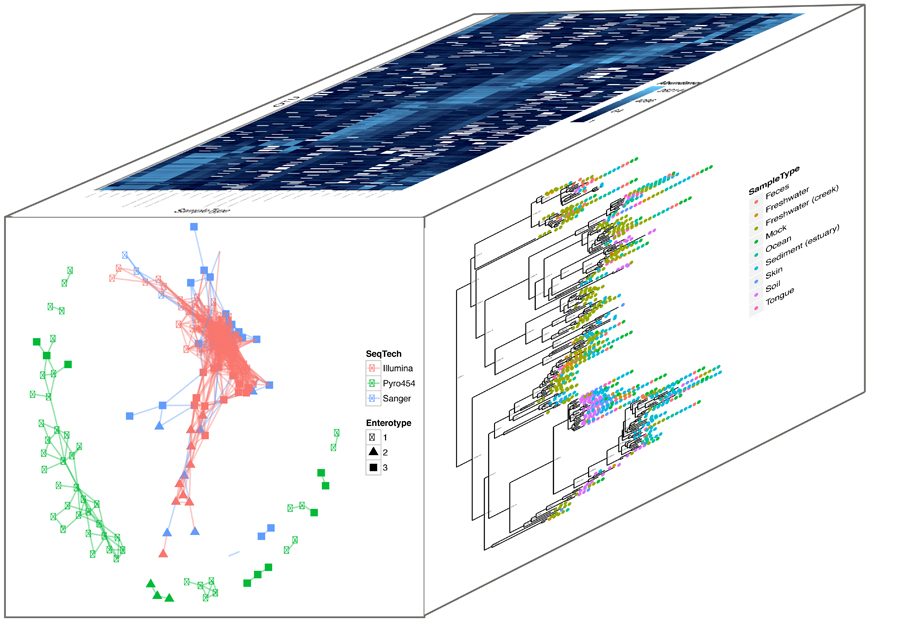 |
| Click to enlarge image. |
| Multi-dimensional aspects of metagenomic data illustrating evolutionary relationships, abundances, and a community network. (Bioconductor package Phyloseq (Joey McMurdie and Susan Holmes). |
The growing complexity and amount of genomic and related data are necessitating novel methods for data synthesis and analysis to answer previously inconceivable questions about biological processes. The Bioinformatics program will be devoted to statistical and mathematical challenges arising in the analysis of such data with the goal of addressing relevant biological questions.
In the planning stages, the structure of the program will revolve around at least the following general topics:
- Statistical pre-processing of emerging high throughput data
- Dependence in high-dimensional data, in particular, high dimensional discrete counts
- Integration of multi-omics data
- Modeling dynamics of mixtures; eg. populations of cells, variants, metagenomics
- Big data and machine learning for addressing 'omic' issues
Tentative list of working groups and leaders:
1. Simultaneous Inference, multiple hypothesis testing -- Fred Wright
2. Dependence in evolutionary models -- Scott Schmidler, Jeff Thorne, Seth Sullivant
3. Dynamics of 3-dim architecture in gene regulation -- Steve Qin, Shili Lin
4. Analysis of high dimensional discrete data -- Dan Nettleton, Fei Zou, David Dunson
5. Sparse Multiway data integration -- Susan Holmes, David Dunson
6. Graphical Modeling for data integration -- Katerina Kechris, Hongyu Zhao
7. Modeling dynamics of communities - metagenomics -- Alex Alekseyeko and Nick Hengartner
8. Imaging genetics -- Hongtu Zhu, Fei Zou
9. Modeling variation in metagenomic networks -- Anthea Monod and Sayan Mukherjee
10. Discovering Complex Human Health Patterns in Multi-omic Data -- J.Paul Brooks and David Edwards
11. Next Generation Sequencing Errors: Xinping Cui and Karin Dorman
Please note that this list is not exhaustive; if you are interested in suggesting and leading a new working group, please email [email protected]
Further Information
For additional information about the program, send e-mail to [email protected]
Workshops
Working Groups
- Spring Course 2014-15: Statistical Learning from Omics Data
- Bio: Data Integration - COPD
- Bio: Next Generation Sequencing Errors
- Bio: Microbiome Community Dynamics and Complexity
- Bio: Analysis of Metagenomic Networks
- Bio: Imaging Genetics
- Bio: Discovering Complex Human Health Patterns in Microbiome Data
- Bio: Data Integration - TCGA
- Bio: Sparse Multiway Data Integration
- Bio: Analysis Of High Dimensional Discrete Data
- Bio: Epigenetics
- Bio: Dependence in Evolutionary Models
- Bio: Multiple Hypothesis Testing and Simultaneous Inference
- Fall Course 2014-15: Statistical and Mathematical Challenges in Molecular Evolution
